Researchers Capture Never-Before-Seen View of Gene Transcription
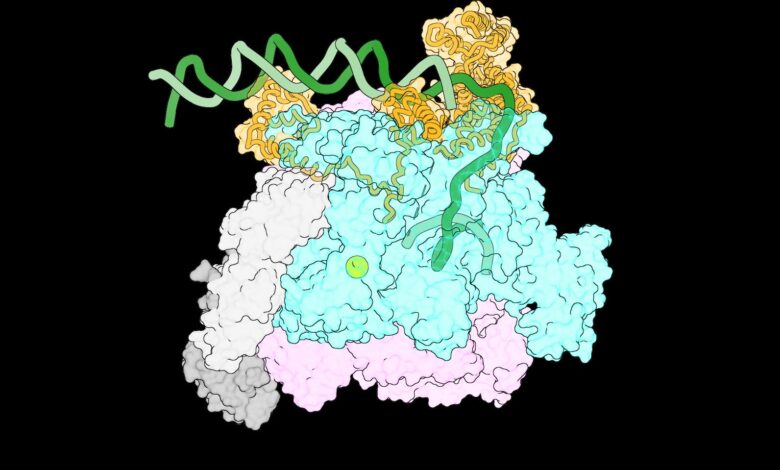
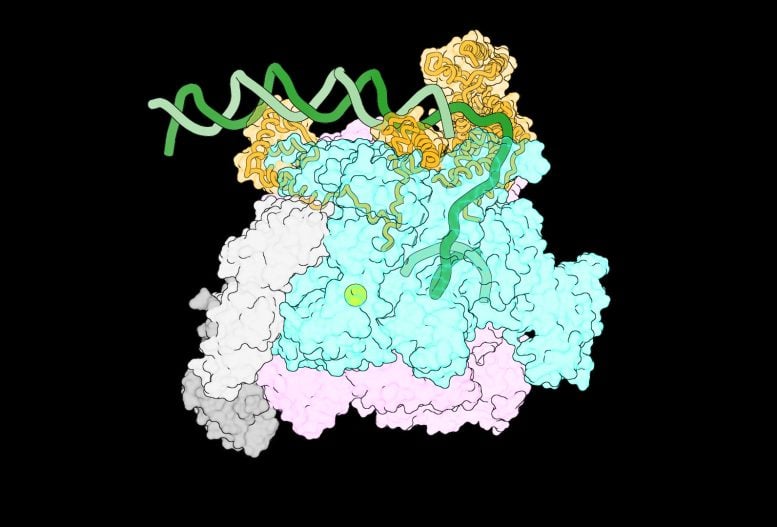
One of the first-ever images of the intermediate complexes that form when RNA polymerase encounters DNA. Credit: Laboratory of Molecular Biophysics at The Rockefeller University
Recent findings illustrate how RNA polymerase interacts with DNA to initiate transcription, captured in milliseconds using advanced microscopy techniques. This breakthrough provides new insights into the mechanisms regulating gene expression, helping resolve long-standing debates in the field.
Every living cell transcribes DNA into RNA. This process starts when an enzyme called RNA polymerase (RNAP) attaches to the DNA. Within a few hundred milliseconds, the DNA double helix unwinds, forming a transcription bubble, allowing one exposed DNA strand to be copied into a complementary RNA strand.
How RNAP accomplishes this feat is largely unknown. A snapshot of RNAP in the act of opening that bubble would provide a wealth of information, but the process happens too quickly for current technology to easily capture visualizations of these structures. Now, a new study in Nature Structural & Molecular Biology describes E. coli RNAP in the act of opening the transcription bubble.
The findings, captured within 500 milliseconds of RNAP mixing with DNA, shed light on fundamental mechanisms of transcription, and answer long-standing questions about the initiation mechanism and the importance of its various steps. “This is the first time anybody has been able to capture transient transcription complexes as they form in real-time,” says first author Ruth Saecker, a research specialist in Seth Darst‘s laboratory at Rockefeller. “Understanding this process is crucial, as it is a major regulatory step in gene expression.”
An unprecedented view
Darst was the first to describe the structure of bacterial RNAP, and teasing out its finer points has remained a major focus of his lab. While decades of work have established that RNAP binding to a specific sequence of DNA triggers a series of steps that open the bubble, how RNAP separates the strands and positions one strand in its active site remains hotly debated.
Early work in the field suggested that bubble opening acts as a critical slowdown in the process, dictating how quickly RNAP can move onto RNA synthesis. Later results in the field challenged that view, and multiple theories emerged about the nature of this rate-limiting step. “We knew from other biological techniques that, when RNAP first encounters DNA, it makes a bunch of intermediate complexes that are highly regulated,” says coauthor Andreas Mueller, a postdoctoral fellow in the lab. “But this part of the process can happen in less than a second, and we were unable to capture structures on such a short timescale.”
To better understand these intermediate complexes, the team collaborated with colleagues at the New York Structural Biology Center, who developed a robotic, inkjet-based system that could rapidly prepare biological samples for cryo-electron microscopy analysis. Through this partnership, the team captured complexes forming in the first 100 to 500 milliseconds of RNAP meeting DNA, yielding images of four distinct intermediate complexes in enough detail to enable analysis.
For the first time, a clear picture of the structural changes and intermediates that form during the initial stages of RNA polymerase binding to DNA snapped into focus. “The technology was extremely important to this experiment,” Saecker says. “Without the ability to mix DNA and RNAP quickly and capture an image of it in real time, these results don’t exist.”
Getting into position
Upon examining these images, the team managed to outline a sequence of events showing how RNAP interacts with the DNA strands as they separate, at previously unseen levels of detail. As the DNA unwinds, RNAP gradually grips one of the DNA strands to prevent the double helix from coming back together. Each new interaction causes RNAP to change shape, enabling more protein-DNA connections to form. This includes pushing out one part of a protein that blocks DNA from entering RNAP’s active site. A stable transcription bubble is thus formed.
The team proposes that the rate-limiting step in transcription may be the positioning of the DNA template strand within the active site of the RNAP enzyme. This step involves overcoming significant energy barriers and rearranging several components. Future research will aim to confirm this new hypothesis and explore other steps in transcription.
“We only looked at the very earliest steps in this study,” Mueller says. “Next, we’re hoping to look at other complexes, later time points, and additional steps in the transcription cycle.”
Beyond resolving conflicting theories about how DNA strands are captured, these results highlight the value of the new method, which can capture molecular events happening within milliseconds in real time. This technology will enable many more studies of this kind, helping scientists visualize dynamic interactions in biological systems.
“If we want to understand one of the most fundamental processes in life, something that all cells do, we need to understand how its progress and speed are regulated,” says Darst. “Once we know that, we’ll have a much clearer picture of how transcription begins.”
Reference: “Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy” by Ruth M. Saecker, Andreas U. Mueller, Brandon Malone, James Chen, William C. Budell, Venkata P. Dandey, Kashyap Maruthi, Joshua H. Mendez, Nina Molina, Edward T. Eng, Laura Y. Yen, Clinton S. Potter, Bridget Carragher and Seth A. Darst, 31 June 2024, Nature Structural & Molecular Biology.
DOI: 10.1038/s41594-024-01349-9