AI Super-Human Eye Brings Scientists One Step Closer to Understanding the Most Complicated and Mysterious Dimension of Our Existence


Kyushu University researchers introduced QDyeFinder, an AI tool that enhances neuron mapping in the brain by using advanced color-coding and machine-learning, showing potential for broader applications in biology.
Researchers have developed QDyeFinder, an AI pipeline that can untangle and reconstruct the dense neuronal networks of the brain.
The brain is the most complex organ ever created. Its functions are supported by a network of tens of billions of densely packed neurons, with trillions of connections exchanging information and performing calculations. Trying to understand the complexity of the brain can be dizzying. Nevertheless, if we ever hope to understand how the brain works, we need to be able to map neurons and study how they are wired.
Now, publishing in Nature Communications, researchers from Kyushu University have developed a new AI tool, which they call QDyeFinder, that can automatically identify and reconstruct individual neurons from images of the mouse brain. The process involves tagging neurons with a super-multicolor labeling protocol, and then letting the AI automatically identify the neuron’s structure by matching similar color combinations.
Challenges in Neuron Mapping
“One of the biggest challenges in neuroscience is trying to map the brain and its connections. However, because neurons are so densely packed, it’s very difficult and time-consuming to distinguish neurons with their axons and dendrites—the extensions that send and receive information from other neurons—from each other,” explains Professor Takeshi Imai of the Graduate School of Medical Sciences, who led the study. “To put it into perspective axons and dendrites are only about a micrometer thick, that’s 100 times thinner than a standard strand of human hair, and the space between them is smaller.”
One strategy for identifying neurons is to tag the cell with a fluorescent protein of a specific color. Researchers could then trace that color and reconstruct the neuron and its axons. By expanding the range of colors, more neurons could be traced at once. In 2018, Imai and his team developed Tetbow, a system that could brightly color neurons with the three primary colors of light.
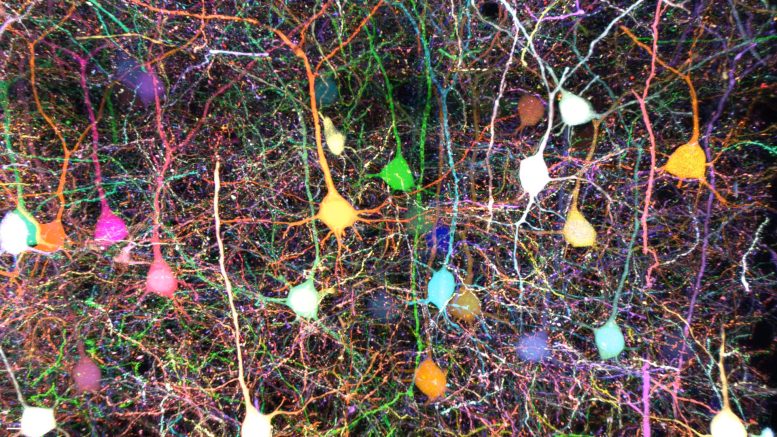
Mouse cortical layer 2/3 pyramidal neurons were labeled with 7-color Tetbow. A combination of 7 fluorescent proteins (mTagBFP2, mTurquoise2, mAmetrine1.1, mNeonGreen, Ypet, mRuby3, tdKatushka2) was used to visualize the dense wiring of neurons. The 7-channel images were then analyzed by the QDyeFinder program to reveal the wiring patterns of individual neurons. Credit: Kyushu University/Takeshi Imai
“An example I like to use is the map of the Tokyo subway lines. The system spans 13 lines, 286 stations, and across over 300 km. On the subway map each line is color-coded, so you can easily identify which stations are connected,” explains Marcus N. Leiwe one of the first authors of the paper and Assistant Professor at the time. “Tetbow made tracing neurons and finding their connections much easier.”
However, two major issues remained. Neurons still had to be meticulously traced by hand, and using only three colors was not enough to decern a larger population of neurons.
Technological Breakthroughs with QDyeFinder
The team worked to scale up the number of colors from three to seven, but the bigger problem then was the limits of human color perception. Look closely at any TV screen and you will see that the pixels are made up of three colors: blue, green, and red. Any color we can perceive is a combination of those three colors, as we have blue, green, and red sensors in our eyes.
“Machines on the other hand don’t have such limitations. Therefore, we worked on developing a tool that could automatically distinguish these vast color combinations,” continues Leiwe. “We also made it so that this tool will automatically stitch together neurons and axons of the same color and reconstruct their structure. We called this system QDyeFinder.”
QDyeFinder works by first automatically identifying fragments of axons and dendrites in a given sample. It then identifies the color information of each fragment. Then, utilizing a machine-learning algorithm the team developed called dCrawler, the color information was grouped together, wherein it would identify axons and dendrites of the same neuron.
“When we compared QDyeFinder’s results to data from manually traced neurons they had about the same accuracy,” Leiwe explains. “Even compared to existing tracing software that makes full use of machine learning, QDyeFinder was able to identify axons with a much higher accuracy.”
The team hopes that their new tool can advance the ongoing quest to map the connections of the brain. They would also like to see if their new method can be applied to the labeling and tracking of other complicated cell types such as cancer cells and immune cells.
“There may come a day when we can read the connections in the brain and understand what they mean or represent for that person. I doubt it will happen in my lifetime, but our work represents a tangible step forward in understanding perhaps the most complicated and mysterious dimension of our existence,” concludes Imai.
Reference: “Automated neuronal reconstruction with super-multicolour Tetbow labelling and threshold-based clustering of colour hues” by Marcus N. Leiwe, Satoshi Fujimoto, Toshikazu Baba, Daichi Moriyasu, Biswanath Saha, Richi Sakaguchi, Shigenori Inagaki and Takeshi Imai, 25 June 2024, Nature Communications.
DOI: 10.1038/s41467-024-49455-y
Funding: Japan Agency for Medical Research and Development, Japan Science and Technology Agency, Japan Society for the Promotion of Science, Uehara Memorial Foundation, Sumitomo Foundation, Ichiro Kanahara foundation, Daiichi Sankyo Foundation of Life Science, Brain Science Foundation